
I am a professor in the Biology Department at the University of Massachusetts, Boston.
I received a BS in Biology from MIT in 1985 and PhD. in Biological Sciences from Stanford University in 1992. From 1992 to 1997, I worked in the Biology Department at MIT; from 1997 to the present, I have been at UMB.
I teach the first semester of the two-semester introductory biology series of courses for Biology Majors (Bio 111). I have flipped this class and developed a wide range of blended learning activities for the course. I discuss the process of flipping the class in a video produced by Nikolai Scherbak. I also teach an upper-level biology course (Bio 362) where students build simulations of biological phenomena using the language StarLogoNova. I have written an on-line textbook for this course which includes sample projects. Finally, I develop educational software and conduct research in Biology and Biology Education. More information on these can be found at the links to the left.
This is an on-line textbook designed to support courses in agent-based simulation using the easy-to-learn drag-and-drop computer language Star Logo Nova. Using materials produced by the MIT Scheller Teacher Education Program, I created the materials to act as a textbook for the "Boot Camp" (learnign to program) portion of project-based simulation courses using this language.
In 2006, Michelle Mishke (MIT) and I published a book of practice problems for Introductory Biology courses. This book is sometimes called “The Problems Book” or “APAIB”. It contains problems on Genetics, Biochemistry, Molecular Biology, and problems that integrate all three components. It also included a set of very detailed solutions to all of the problems in the book. The book also makes extensive use of software simulations for practicing with these materials.
In 2012, the book went out of print. Fortunately, the publisher (ASM Press) was generous enough to allow us to post the content on-line for free. It is available from the links below:
I am part of the BioQUEST Curriculum Consortium and through this I develop software for teaching biology. I work in close collaboration with Ethan Bolker of the UMB CS department and his students to develop much of this software.
These are designed to allow students to explore biological phenomena through interactive simulations.
All are freely-available from the links below, open-source, and subject to the GNU Public License (GPL).
All are written in Java, which is freely-available at this link.
Many also have javascript versions that will run in most browsers (especially Google Chrome). These versions lack some of the features of the stand-alone Java programs, but many allow on-line grading.
If you use this software for teaching, please let me know. That way, I can collect feedback for further development and document uses for grant-development purposes. THANKS!
| Aipotu. An interactive simulation linking Genetics, Biochemistry, Molecular Biology, and Evolution. jsAipotu is a javascript version of Aipotu that runs in a web browser. |  |
| The Virtual Genetics Lab. An interactive simulation of Mendelian Genetics. jsVGL is a javascript version of VGL that runs in a web browser. |  |
| The Gene Explorer. An interactive simulation of gene expression. jsGenex is a javascript version of Genex that runs in a web browser. |  |
| The Protein Investigator. An interactive simulation of protein folding that allows you to fold proteins of your own design. jsPI |  |
| jlogP. Calculates the hydrophobicity and molecular formula of molecules drawn by the user. jsMolCalc is a javascript version of MolCalc that runs in a web browser. |  |
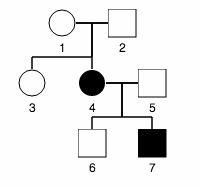
| jsPedigrees. A tool for exploring pedigree genetics. Students can draw a pedigree and then the software will determine which modes of inheritance are compatible. |  |
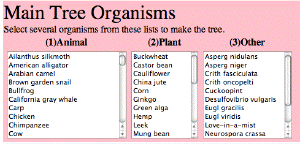
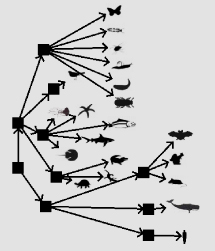
| The Phylogenetic Tree Constructor. A tool that allows students to select organisms for constructing trees using molecular phylogeny software. |  |
| Tree Building Applet. Allows students to draw a phylogenetic tree of 20 familiar organisms. |  |
I have also developed several other pieces of Biology Curriculum.
In 2013, I teamed up with Eric Lander from MIT's Broad Institute to develop a Massively Open On-line Course (MOOC) in General Biology called "7.00x: The Secret of Life". This free on-line course includes lectures, problem sets, exams, and more. It uses the software I have developed in the problem sets and exams. It can be found at edX. The MOOC was integrated into Bio 111 starting in Fall 2013.
My research program investigates Biology Education.
I am currently looking for graduate students to work on these projects as well as others. Interested students should contact me by e-mail brian.white@umb.edu; information on applying to the UMB Biology Graduate Program can be found here.
My research program focuses on the development and evaluation of materials (often, but not always software) for teaching Biology at the undergraduate level.
Our current research focuses on students' use of the on-line materials in the flipped classroom - specifically, my Bio 111 course which I flipped in Fall 2014.
We have extensively analyzed the clickstream log files generated by the on-line materials (called the "SPOC" and hosted by edX). We have explored a variety of questions:
Possible projects include:
Past projects have resulted in the following publications:
I also like to build electronic projects on my spare time. I have a website that describes these projects.